GRAPE
The Automatic Parameter Estimation
Program for

Introduction
GRAPE (GRadient Ascent Parameter Estimation) is the automatic
training program for the ab initio
eukaryotic gene finder GeneZilla. The program uses gradient ascent optimization (also
called "hill-climbing") to fine
tune the parameters of the gene finder, as described in:
Majoros W. and
Salzberg S.L. (2004) An
empirical analysis of training protocols for probabilistic gene
finders, BMC Bioinformatics 5:206.
The procedure is illustrated schematically below:
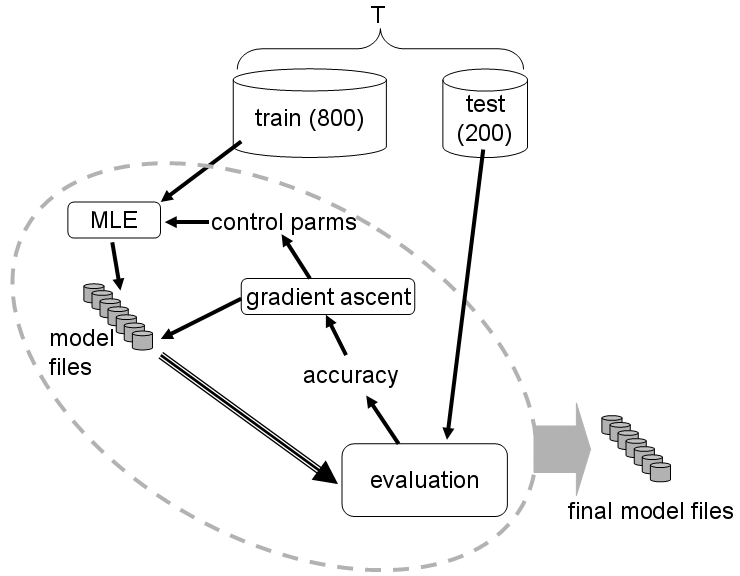
The program works by iteratively refining the current parameters of the
gene finder so as to incrementally improve accuracy on a set of test
genes. The relative sizes of the training and test sets shown in the
figure are suggestive only. Note that the final accuracy measurements
on the test set should not be used for publication purposes, as this
would constitute a form of post hoc
prediction, and is therefore generally invalid for objective accuracy
assessment.
The GRAPE program, being a research tool, is continually undergoing
modifications. Currently, the program is designed to optimize
the following parameters:
- mean intron, intergenic, and UTR lengths
- transition probabilities
- exon "optimism"
- signal sensor window sizes
- signal sensor Markov order
- signal sensitivity
- number of signal boosting iterations
Using GRAPE to optimize other parameters than these requires
modifications to the GRAPE source code. GRAPE is written in Perl
and is provided as open-source software. Modifications are
welcome, as are contributions and enhancements to the continually
evolving code base of the GeneZilla and GRAPE projects. Contact bmajoros@tigr.org for information
on contributing to the software development of these projects.
GRAPE is currently configured to process only a single isochore at a
time. If you wish to use the gene finder in multiple-isochore
mode, you will have to apply GRAPE to each isochore separately; it is
recommended that this be done in a separate directory for each isochore.
The usage statement for the program is:
GRAPE.pl
<logfile.txt> <max-num-test-genes>
<genome-GC-content> [-d]
where -d = optimize exon duration distributions
<genomic-GC-content> is between 0 and 1 (ex: 0.48)
Prerequisites:
1) Must have training set
in ./iso0-100.gff
2) Must have test set in
./test.gff
3) Must have contigs in
./iso0-100.fasta
4) Run get-examples.pl
iso0-100.gff iso0-100.fasta TAG,TGA,TAA notrim
5) Run
get-duration-distr.pl for each type of exon
(and
view results using xgraph to ensure smoothness)
The <logfile.txt>
file will be overwritten by the program to contain a record of the
optimization steps performed during hill-climbing. The <max-num-test-genes>
places a limit on the number of test genes in order to accelerate the
evaluation step of the hill-climbing procedure. <genome-GC-content>
must be a real number between 0 and 1. The prerequisites for
running the program are given above. Information on performing these
prerequisite steps can be found here.
A number of additional parameters are defined within the GRAPE program:
my $MIN_SAMPLE_SIZE=175;
my $EXON_POOLING_THRESHOLD=50;#
we pool if fewer than this
my
$FORCE_EXON_POOLING=1; # always pool exons?
my
$MAX_BWM_SAMPLE_SIZE=40; # use BWM if <40, otherwise use
WAM
my
$MIN_NONCODING_MARGIN=10; # min size for noncoding signal margin
my
$MAX_NONCODING_MARGIN=45; # max size for noncoding signal margin
my
$MIN_CODING_MARGIN=0; # min size for
coding signal margin
my
$MAX_CODING_MARGIN=10; # max size for coding
signal margin
BWM denotes a Binomial Weight Matrix,
which utilizes a binomial test to decide whether to use background
positional nucleotide frequencies in the presence of small sample sizes.
The GRAPE.pl
program may take several hours to complete, depending on the hardware
on which it is executed. The result of running the program will
be a grape.iso
file and a set of corresponding *.cfg files and model
files having extensions *.model
and *.trans.
The grape.iso
file can be used directly by the GeneZilla program, though
modifications to the path names may be necessary before deployment on
another filesystem.

